Physical Model Maps Cas9 Cutting Behavior
March 23, 2022| |
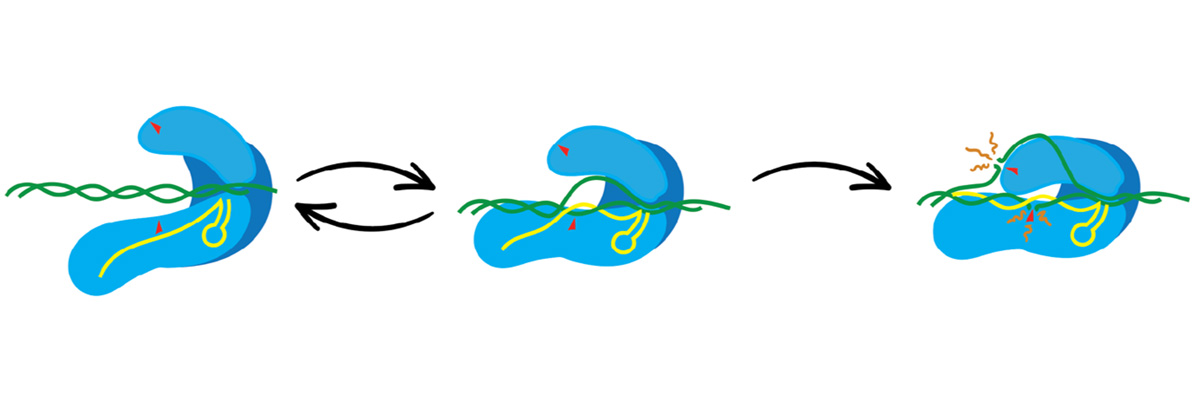
Researchers from Delft University of Technology (TU Delft) have developed a physical-based model that establishes a quantitative framework on how CRISPR-Cas9 works, and allows them to predict where, with what probability, and why targeting errors (off-targets) occur.
A research team led by Martin Depken at TU Delft's Department of Bionanoscience shows how the new, physical-based model greatly improves existing models not only by predicting where the DNA is likely to be cut but also with what probability this will happen. Depken's team used a physics-based approach.
“In gene editing, you want to maximize the probability of cutting at the intended site, while minimizing the amount of cutting in the rest of the genome," Depken said. They created a model that can do this. He added that their model "changes the way in which to describe the gene editing from a binary choice to a complete probabilistic picture."
For more details, read the article on the TU Delft website.
| |
You might also like:
- Redesigned Cas9 Makes Gene Editing Safer Without Sacrificing Speed
- Cas-CLOVER, an Alternative to CRISPR-Cas9
- Pocket K No. 54: Plant Breeding Innovation: CRISPR-Cas9
Biotech Updates is a weekly newsletter of ISAAA, a not-for-profit organization. It is distributed for free to over 22,000 subscribers worldwide to inform them about the key developments in biosciences, especially in biotechnology. Your support will help us in our mission to feed the world with knowledge. You can help by donating as little as $10.
-
See more articles:
-
Gene Editing Supplement (March 23, 2022)
-
Research and Tools
- Spineless Fish Developed Through Genome Editing in China
- Physical Model Maps Cas9 Cutting Behavior
- Experts Develop PiggyBac for Delivery of CRISPR Tools into Stem Cells
-
Policy Considerations and Approvals
- Swiss Parliament Eases Restriction for Gene Editing
- NIAB Lauds UK Parliamentary Approval of New Gene Editing Rules
-
Trends and Impact
- Genome Editing Can Help Achieve UN Sustainable Development Goals
-
Read the latest: - Biotech Updates (February 11, 2026)
- Gene Editing Supplement (January 28, 2026)
- Gene Drive Supplement (February 22, 2023)
-
Subscribe to BU: - Share
- Tweet