Global Consortium of Scientists Develop Affordable Sequencing Method for Pathogen Analysis to Help Tackle Global Epidemics
January 5, 2022| |
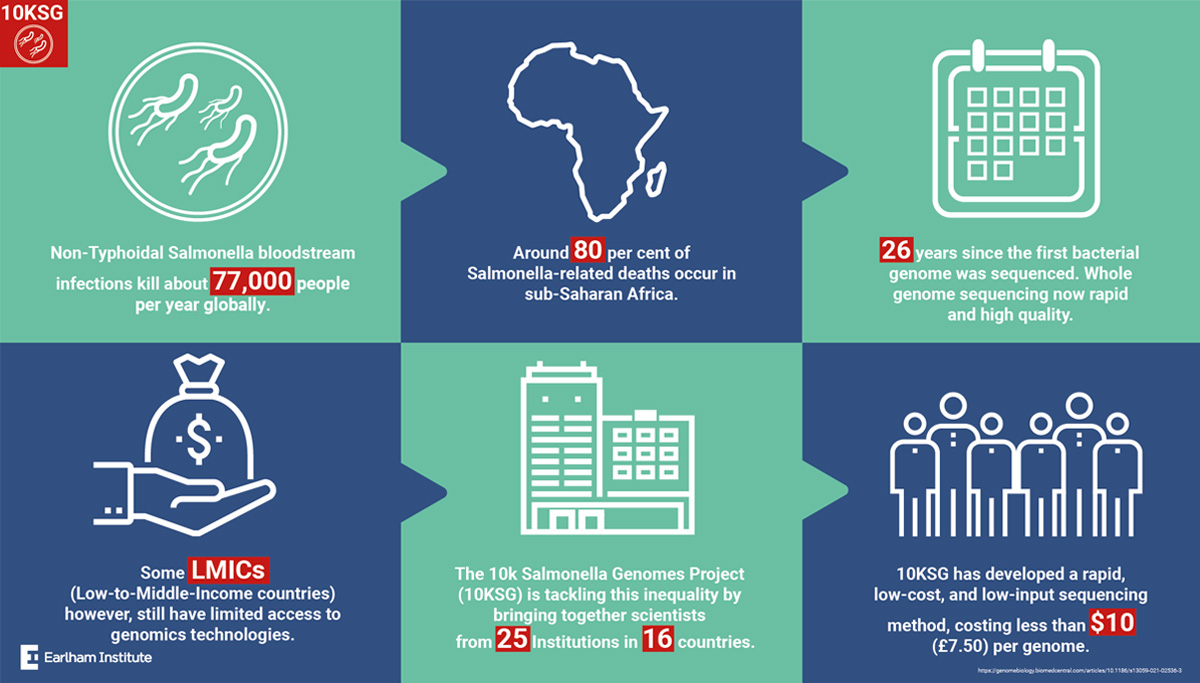
A global consortium of scientists led by Earlham Institute and the University of Liverpool in the United Kingdom marked a significant milestone by developing cheap and accessible methods for sequencing large collections of bacterial pathogens that low- and middle-income countries (LMICs) can use at a cost of less than US$10 per genome.
During the coronavirus pandemic, genomic surveillance has been in the spotlight and the ability of countries to contribute through low-cost and rapid whole genome sequencing (WGS) has become increasingly important. The methods developed by the consortium can be applied to large collections of pathogens and will strengthen global research collaborations to tackle future pandemics.
Focusing on the pathogen Salmonella enterica, this large-scale genomic sequencing initiative was led by the worldwide 10,000 Salmonella genomes research consortium (10KSG) with scientists from 16 countries. 10KSG aims to make genomic data more accessible to LMICs and the project has sequenced and analyzed 10,000 Salmonella genomes from Africa and Latin America. The researcher's innovative WGS approach aimed to streamline the large-scale collection and genome sequencing of bacterial isolates and collected more than 10,400 clinical and environmental bacterial isolates from LMICs in under a year.
The sample logistics pipeline, developed by the University of Liverpool, was optimized by shipping the heat-inactivated bacterial isolates as ‘thermolysates' in ambient conditions from different countries to the UK. The isolates were sequenced at the Earlham Institute using the unique LITE protocol - a low-cost, low input automated method for rapid genome sequencing. In total, the gene library construction and DNA sequencing bioinformatic analysis were done with a total reagent cost of less than US$10 (around £7.50GBP) per genome.
For more details, read the article in the Earlham Institute Newsroom.
| |
You might also like:
- ICRISAT Leads Largest Plant Genome Sequencing Effort, Yields Chickpea Pan-Genome
- Genome Sequencing of 445 Varieties Reveals Domestication History of Cultivated Lettuce
- Endangered Plant Species Becomes Model for Genome Sequencing
Biotech Updates is a weekly newsletter of ISAAA, a not-for-profit organization. It is distributed for free to over 22,000 subscribers worldwide to inform them about the key developments in biosciences, especially in biotechnology. Your support will help us in our mission to feed the world with knowledge. You can help by donating as little as $10.
-
See more articles:
-
News from Around the World
- Global Consortium of Scientists Develop Affordable Sequencing Method for Pathogen Analysis to Help Tackle Global Epidemics
- Experts: Allow Golden Rice to Save Lives
- UCR Scientists Breeding HLB Tolerant Citrus
- Political Will and Scientific Community Continue to Move Biotech Adoption in Bangladesh
- Australian OGTR Receives Application for GM Canola and Indian Mustard Field Trial
- GM Soybean and GM Maize Industrialization in China Shows Remarkable Results
- Göttingen University Research Team Develops Method to Complete Genetic Data
- High-Iron Wheat Awaits Approval for Field Trials in the UK
-
Research Highlights
- International Research Team's Genetic Discoveries to Improve Spinach Disease Resistance and Palatability
- Lychee's Genome Unveils Its Domestication History
-
Plant
- CRISPR-Cas9 Reveals Role of SlSRM1-like in Leaf Development
- CRISPR Continues to Drive Gene Editing Market Growth, Report
-
Read the latest: - Biotech Updates (December 17, 2025)
- Gene Editing Supplement (December 17, 2025)
- Gene Drive Supplement (February 22, 2023)
-
Subscribe to BU: - Share
- Tweet