Pocket K No. 36: Marker-Free GM Plants
| |
Introduction
Selectable marker genes are vital to the research and development of genetically modified (GM) crops. The methods used to introduce foreign DNA in a plant cell, either by microinjection, particle gun, electroporation or Agrobacterium, are relatively inefficient. Pinpointing cells that successfully incorporated foreign DNA in an ocean of non-transformed cells is akin to finding a needle in a haystack. To find transgenic cells, a marker gene is co-introduced with the gene of interest. These dominant genes confer resistance to antibiotics, such as hygromycin (hpt) and kanamycin (nptII), and herbicides, such as phosphinothricin (bar) and chlorosulfuron (als), that kill non-transformed cells. However, antibiotic and herbicide resistance marker genes may not be required in mature plants, especially when they are cultivated in fields.
 The presence of these marker genes in commercialized transgenic crops has caused considerable public concern about the medical implications of GM food consumption and GM crop cultivation. Herbicide resistance genes might be transferred by outcrossing to weeds and wild crop relatives. There also exists the possibility, albeit extremely rare, of horizontal gene transfer from transgenic plants to soil and intestinal microorganisms, resulting in pathogens against which antibiotics currently being used are rendered ineffective. However, to date, no experiment has provided any evidence that the antibiotic markers presently in use pose risks to human or animal health.
The presence of these marker genes in commercialized transgenic crops has caused considerable public concern about the medical implications of GM food consumption and GM crop cultivation. Herbicide resistance genes might be transferred by outcrossing to weeds and wild crop relatives. There also exists the possibility, albeit extremely rare, of horizontal gene transfer from transgenic plants to soil and intestinal microorganisms, resulting in pathogens against which antibiotics currently being used are rendered ineffective. However, to date, no experiment has provided any evidence that the antibiotic markers presently in use pose risks to human or animal health.
Not all scientists agree with these claims. The difficulty of proving that the marker genes are indeed harmless has significantly limited the public acceptance of agricultural biotechnology.
 A lot of research effort has been directed towards the development of marker-free transformation methods and selectable marker elimination strategies. Besides minimizing public concerns, the absence of resistance genes in transgenic plants could also reduce the costs for developing GM products and lessen the need for time-consuming safety evaluations, thereby speeding up the commercial release of new products. Generation of marker free plants likewise supports single line re-transformation, an important approach towards introduction of multiple genes for complex traits such as resistance to several pathogens and tolerance to abiotic stress.
A lot of research effort has been directed towards the development of marker-free transformation methods and selectable marker elimination strategies. Besides minimizing public concerns, the absence of resistance genes in transgenic plants could also reduce the costs for developing GM products and lessen the need for time-consuming safety evaluations, thereby speeding up the commercial release of new products. Generation of marker free plants likewise supports single line re-transformation, an important approach towards introduction of multiple genes for complex traits such as resistance to several pathogens and tolerance to abiotic stress.
There are several ways to either avoid or get rid of selectable marker genes. Methods that will allow the removal of DNA in plants as efficiently as it is inserted have been developed, such as the use of site-specific recombination, transposition and homologous recombination. Researchers have also described several substitute marker genes that have no harmful biological activities. The presence of these non-bacterial genes allows the plants to metabolize non-toxic agents normally harmful to them.
Alternatives to Antibiotic/Herbicide Resistance Markers
 Scientists have identified positive selectable marker genes that are dependent on non-toxic substances that may be substrates for growth or that induce growth of transformed cells or tissues. These selectable markers only suppress the growth of non-transformed cells, in contrast to antibiotic and herbicide resistance markers.
Scientists have identified positive selectable marker genes that are dependent on non-toxic substances that may be substrates for growth or that induce growth of transformed cells or tissues. These selectable markers only suppress the growth of non-transformed cells, in contrast to antibiotic and herbicide resistance markers.
For instance, transgenic events can be selected using markers that enable them to use a particular food source. An example of this approach is the use of phosphomannose isomerase gene (pmi). Cells that successfully incorporated foreign DNA can be identified since they are capable of utilizing mannose as their sole food source. PMI has been used as a selectable marker for transformation of many plant species, including rice, wheat, millet, tobacco, sugarcane, apple and onion. Other selection systems that rely on the growth of plant cells in the presence of a particular sugar or sugar alcohol as their sole energy source include the use of xylA (xylose), atlD (arabitol) and AtTPS1 (glucose). Genes that allow plants to survive in media supplied with amino acid analogs and D-amino acids, such as TSB1 and ASA2 (tryptophan), TD (isoleucine), and dao1 (D-amino acids), have also been used to identify transformed cells.
The use of alternative markers completely eliminates concerns over the possible spread of antibiotic and herbicide resistance genes into the environment. However, since these markers entail the introduction of new metabolic pathways a more rigorous risk assessment will be needed to establish the safety of the resulting transgenic products.
Researchers have also used visible markers to make transgenic plants visually recognizable. The jellyfish gene for the green florescent protein (GFP) makes genetically modified plant cells appear green when exposed to ultraviolet light. Reporter genes such as the firefly protein luciferase and plants’ red-purple anthocyanins have likewise been used as visible markers for selecting stably transformed cell lineages. A major disadvantage of this approach, however, is that transformed and non-transformed cells must later be manually separated, which can be very tedious and time consuming.
Conventional Genetics Lends A Hand: Co-transformation
Although numerous alternative markers exist, complete removal of selectable markers might be more favorable in the long run since it is very likely that future regulatory legislation will strongly favor the absence of superfluous transgenic material in GM crops.
One of the simplest marker removal strategies is the co-transformation approach. The principle of the strategy is the integration of the transgene of interest and the marker gene into different unlinked locations in the plant genome and their subsequent segregation in the next generation to yield progenies carrying the transgene but not the markers. Three approaches are used for co-transformation: (i) introducing two different Agrobacterium strains each with a transformation vector, one carrying the marker gene and the other the target gene, (ii) using one bacterial strain carrying two vectors, each with one gene, and (iii) using a bacterial strain harboring one vector, with the two genes at separate sites.
Co-transformation can be readily integrated into existing transformation protocols. It has been used to successfully eliminate selectable marker genes in several crop plants, with transformation frequencies reaching as high as 85%. However, since the approach relies on the segregation of genes during sexual reproduction, it cannot be used for vegetatively propagated plants. Selection of the progenies carrying only the target gene may also prove to be laborious, since statistically the desired trait combination can only be found in one out of 16 progenies.
Molecular Cut and Paste: Site Specific Recombination
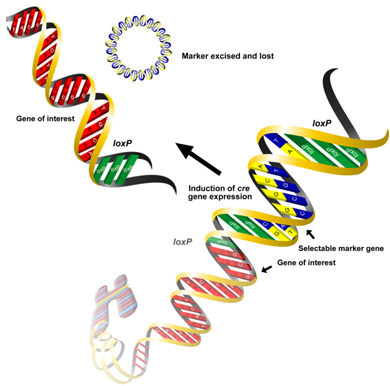 Microbial site specific recombinases have also been used to eliminate unwanted markers from GM plants. These enzymes act as molecular scissors capable of cleaving DNA at specific sites. They can also act as molecular glue, ligating the cleaved DNA fragments at a second target sequence. The gene encoding these enzymes is introduced along with the marker gene, which is flanked by palindromic sequences recognizable by the enzyme, and the gene of interest. Once transformed cells have been selected, the recombinase gene can be activated by an external stimulus. The recombinases then cut out the marker genes and the genes for the enzymes themselves, making the resulting plants devoid of any selectable markers.
Microbial site specific recombinases have also been used to eliminate unwanted markers from GM plants. These enzymes act as molecular scissors capable of cleaving DNA at specific sites. They can also act as molecular glue, ligating the cleaved DNA fragments at a second target sequence. The gene encoding these enzymes is introduced along with the marker gene, which is flanked by palindromic sequences recognizable by the enzyme, and the gene of interest. Once transformed cells have been selected, the recombinase gene can be activated by an external stimulus. The recombinases then cut out the marker genes and the genes for the enzymes themselves, making the resulting plants devoid of any selectable markers.
There are three well described site-specific recombination systems that have been used successfully for the removal of marker genes, the most widely used of which is the Cre/loxP system from the bacteriophage P1 (see Figure). The Cre recombinase catalyzes a reaction between two loxP sequences and results in excision of the DNA fragment between them. The Cre recombinase gene can be introduced into transgenic plants either by re-transformation, breeding or inducible autoexcision. The autoexcision strategy is a one-step process that relies on chemically-inducible promoters for gene activation. Several experiments have demonstrated the advantages of this method in comparison to re-transformation and crossing.
The lysine-fortified transgenic maize LY038, in which the marker gene has been removed with the help of the Cre-lox system, has been approved for cultivation in Canada, Japan and the U.S. and for food and feed use in Australia, Mexico and the Philippines.
Jumping Along Chromosomes: Marker Deletion via Transposons
The process that enables certain genes to ‘jump’ at a certain position on the plant genome can also be used to generate marker-free plants. The approach is analogous to site-specific recombination, only that instead of a recombinase and recognition sites, transposons or jumping genes are used. Transposons contain a gene for a special enzyme, which recognizes certain signals in the DNA. The enzyme cuts the DNA fragment flanked by these signals and integrates them randomly in the genome. The most characterized transposons are those of the Ac/Ds family, which was first discovered in maize, the special enzyme being the Ac (activator) transposase and the Ds (dissociator) sequences the tag signals.
The gene of interest or the marker gene can be placed within the ‘jumping’ sequence, in such a way that the two genes can be separated from each other upon the activation of transposase. Although the system has been shown to be effective, marker removal efficiency via this strategy is poor, due to the low incidence of occurrence. This approach can also be time-consuming since breeding or segregation is required to separate the gene of interest and the marker gene.
Future Prospects
Numerous approaches to eliminate antibiotic and herbicide markers have been developed over the last several years and further improvements are now underway. Recently, researchers have described procedures to eliminate residual recognition sequences at recombination sites. This may increase the appeal of site specific recombination as the tool of choice to remove unwanted DNA sequences. Scientists are also searching for ways to hasten the selection of marker-less progenies after co-transformation or transposition. Novel marker elimination strategies based on gene targeting and homologous recombination have been reported. With these developments, the concern about an uncontrolled spread of antibiotic and herbicide resistance genes in the environment might become irrelevant in the future.
Advances on the use of zinc-finger nucleases have also been reported and their potential in removing transgenes and utility in targeted gene replacement offer much promise.
| |
References
- Hare PD and Chua NH. (2002) Nat. Biotechnol. 20:113-122
- Sundar IK and Sakthivel N. (2008) J Plant Physiol 165: 1698-1716
- Kondrák M. et al. (2007) ISB News Report
- http://www.isb.vt.edu/news/2007/news07.apr.htm#apr0705
- Tian L. (2007) ISB News Report http://www.isb.vt.edu/news/2007/news07.jul.htm#jul0701
- Darbani B. et al. (2007) Biotechnol. J. 2:83–90
- Puchta H. (2003) Plant Cell, Tissue and Organ Culture 74:123-134
- Verweire D et al. (2007) Plant Physiology 145:1220-1231
- Miki B and McHugh S. (2004) Journal of Biotechnology 107:93-232
- Ramessar K et al. (2007) Transgenic Research 16:261-280
- Marker Gene Elimination, Concepts and Research Projects, GMO Safety http://www.gmosafety.eu/en/gene_transfer/elimination/
- Alternatives to Antibiotic Resistance Markers, GMO Compass
http://www.gmo-compass.org/eng/safety/human_health/
129.alternatives_antibiotic_resistance_marker_genes.html - Goldstein DA et al. (2005) J. Applied Microbiology 99:7-23
- Keenan RJ and Stemmer WPC. (2002) Nat. Biotechnol. 20:215-216
- Lloyd, A et al. (2005) PNAS 102:2232-2237
- Stewart, CN et al. (2009) http://www.reeis.usda.gov/web/crisprojectpages/207870.html
