Pocket K No. 19: Molecular Breeding and Marker-Assisted Selection
| |
The process of developing new crop varieties can take almost 25 years. Now, however, biotechnology has considerably shortened the time to 7-10 years for new crop varieties to be brought to the market. One of the tools which can make it easier and faster for scientists to select plant traits is marker-assisted selection (MAS).
Molecular shortcut
|
The differences that distinguish one plant from another are encoded in the plant’s genetic material, the DNA. DNA is packaged in chromosome pairs (strands of genetic material), one coming from each parent. The genes, which control a plant’s characteristics, are located on specific segments of each chromosome. Together, all of a plant’s genes make up its genome.
Some traits, like flower color, may be controlled by only one gene. Other more complex characteristics, however, like crop yield or starch content, may be influenced by many genes. Traditionally, plant breeders have selected plants based on their visible or measurable traits, called the phenotype. This process can be difficult, slow, influenced by the environment, and costly – not only in the development itself, but also for the economy, as farmers suffer crop losses.
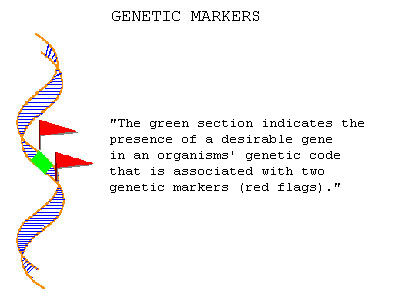
As a shortcut, plant breeders now use marker-assisted selection (MAS). To help identify specific genes, scientists use what are called molecular or genetic markers. The markers are a string or sequence of nucleic acid which makes up a segment of DNA. The markers are located near the DNA sequence of the desired gene and are transmitted by the standard laws of inheritance from one generation to the next (Figure 1). Since the markers and the genes are close together on the same chromosome, they tend to stay together as each generation of plants is produced. This is called genetic linkage. This linkage helps scientists to predict whether a plant will have a desired gene. If researchers can find the marker for the gene, it means the desired gene itself is present.
As scientists learn where markers occur on a chromosome, and how close they are to specific genes, they can create a genetic linkage map. Such a map would show the location of markers and genes, and their distance from other known genes. Scientists can produce detailed maps in only one generation of plant breeding.
Using very detailed genetic maps and better knowledge of the molecular structure of a plant’s DNA, researchers can analyze only a tiny bit of plant tissue, even from a newly germinated seedling. Once the tissue is analyzed, scientists know whether that seedling contains the appropriate gene. If it doesn’t, they can quickly move on and concentrate on analysis of another seedling, eventually working only with the plants which contain a specific trait.
It should be noted, however, that molecular breeding through MAS is somewhat limited in scope compared to genetic engineering or modification because: 1) it works only for traits already present in a crop; 2) it cannot be used effectively to breed crops which have long generation times (e.g. citrus); and 3) it cannot be used effectively with crops which are clonally propagated because they are sterile or do not breed true (this includes many staples such as yams, bananas, plantain, sweet potato, and cassava).
Molecular Markers
Several marker systems have been developed and are applied to a range of crop species. These are the Restriction Fragment Length Polymorphisms (RFLPs), Random Amplification of Polymorphic DNAs (RAPDs), Sequence Tagged Sites (STS), Amplified Fragment Length Polymorphisms (AFLPs), Simple Sequence Repeats (SSRs) or microsatellites, and Single Nucleotide Polymorphism (SNPs). The advantages and disadvantages of these marker systems are provided in Table 1.
| Table 1: Comparison of most commonly used marker systems (adopted from Korzun, 2003) | |||||
| Feature | RFLPs | RAPDs | AFLPs | SSRs | SNPs |
| DNA required (?g) | 10 | 0.02 | 0.5-1.0 | 0.05 | 0.05 |
| DNA quality | High | High | Moderate | Moderate | High |
| PCR-based | No | Yes | Yes | Yes | Yes |
| Number of polymorph loci analyzed | 1.0-3.0 | 1.5-50 | 20-100 | 1.0-3.0 | 1.0 |
| Ease of use | Not easy | Easy | Easy | Easy | Easy |
| Amenable to automation | Low | Moderate | Moderate | High | High |
| Reproducibility | High | Unreliable | High | High | High |
| Development cost | Low | Low | Moderate | High | High |
| Cost per analysis | High | Low | Moderate | Low | Low |
These molecular techniques have been widely used to monitor differences in DNA sequence in and among species. They also allow the creation of new sources of genetic variation by introducing new and desirable traits from wild varieties into elite lines. While RFLP markers have been the basis for most genetic work in crop plants, AFLPs and SSRs are currently the most popular techniques used due to ease in detection and automation. The adoption of the new marker system, SNPs, is now highly preferred, with the increasing amount of sequence information, and the determination of gene function due to genomic research.
|
|
The main uses of these molecular markers in crop genetic studies are as follows:
- Assessment of genetic variability and characterization of germplasm
- Identification and fingerprinting of genotypes
- Estimation of genetic distances between population, inbreeds, and breeding materials
- Detection of monogenic and quantitative trait loci (QTL)
- Marker-assisted selection
- Identification of sequences of useful candidate genes
MAS for Pathogen Resistance in Tomato
|
One of the major constraints in tomato cultivation and production are severe harvest losses caused by a number of pathogens, including viruses, bacteria, fungi, and nematodes. Farmers have adopted control measures, such as applications of agrochemicals and use of resistant lines. Although conventional breeding has had a significant impact on improving resistance of tomato, the time-consuming process of making crosses and backcrosses, and the selection of the desired resistant progeny make it difficult to react adequately to the evolution of new virulent pathogens.
Molecular markers are now being widely used for breeding tomato. More than 40 genes that confer resistance to major classes of tomato pathogens have been mapped, cloned, and/or sequenced (Grube, et. al., 2000). These maps have allowed for “pyramiding” resistance genes in tomato through MAS, where several resistance genes can be engineered into one genotype. Currently, tomato breeding through MAS has resulted in varieties with resistance or tolerance to one or more specific pathogens.
Glossary
AFLP: Amplified Fragment Length Polymorphism. A highly sensitive method for detecting DNA polymorphism. Following restriction enzyme digestion of DNA, a subset of the DNA fragments is selected for PCR amplification and visualization.
Genetic Map: A map of the relative positions of genetic loci on a chromosome, determined on the basis of how often the loci are inherited together.
Linkage Map: A map of relative positions of genes on a chromosome. Genes inherited together are close to each other on the chromosome, and said to be linked.
Microsatellites: Very short DNA motifs (1-10 base pairs) which occur as tandem repeats at numerous loci throughout the genome. Also known as simple sequence repeats (SSR), simple tandem repeats or simple repetitive sequences.
Monogenic trait (Mendelian trait): a trait determined by the action of a single genetic locus
Nucleic acid: molecule found in all living cells, in which the hereditary information is stored and from which it can be transferred. The two chief types are DNA (deoxyribonucleic acid), found mainly in cell nuclei, and RNA (ribonucleic acid), found mostly in cytoplasm.
PCR: Polymerase Chain Reaction. A method for amplifying a DNA sequence in large amounts using a heat-stable polymerase and suitable primers to direct the amplification of the desired region of DNA.
Polymorphism: A detectable difference at a particular gene or marker occurring among individuals.
RAPD: Random Amplification of Polymorphic DNA. A widely-used technique for amplifying anonymous stretches of DNA using PCR with arbitrary primers.
RFLP: Restriction Fragment Length Polymorphism. Variations which occur in the length of DNA fragments produced when DNA is broken down by restriction enzymes (enzymes which recognize specific sequences of DNA, usually 4-6 base pairs long, and cleave the DNA at these points, known as restriction sites).
SNP: Single Nucleotide Polymorphism. A common, but minute, variation that occurs in DNA sequences of a genome. These
variations can be used to track inheritance in families or species.
QTL: Quantitative Trait Locus. Location of a specific gene that affects a measurable or quantifiable trait. These traits are typically affected by more than one gene, and also by the environment. Examples of quantitative traits are plant height (measured on a ruler) and body weight (measured on a balance)
Quantitative (continuous) traits: phenotypes that exhibit a range of measurable outcomes.
| |
References:
- Ag-West Biotech Inc. 1998. Marker assisted selection: Fast track to new crop varieties. Agbiotech Infosource. Canada. (http://www.agwest.sk.ca/sabic_bioinfo.shtml)
- Barone, A. 2003. Molecular marker-assisted selection for resistance to pathogens in tomato. A paper presented during theFAO international workshop on “Marker assisted selection: A fast track to increase genetic gain in plant and animal breeding?”. 17-18 October 2003, Turin, Italy.
- FAO 2002 Crop Biotechnology: A working paper for administrators and policy makers in sub-Saharan Africa. Kitch, L., Koch, M., and Sithole-Nang, I.
- Grube, R.C., Radwanski, E.r., Jahn, M. 2000. Comparative genetics of disease resistance within the Solanaceae. Genetics 155: 873-887.
- Korzun, V. 2003. Molecular markers and their applications in cereals breeding. A paper presented during the FAO international workshop on “Marker assisted selection: A fast track to increase genetic gain in plant and animal breeding?”. 17-18 October 2003, Turin, Italy.
- http://www.ipgri.cgiar.org/training/unit10-1-4/Glossary.pdf
- http://www.ostp.gov/html/plantgenome/abstract.html
- http://usda-ars-beaumont.tamu.edu/dblhelix.jpg